The Time-series Explorer is a novel information visualisation technique using animated views to support the exploratory analysis of microarray time-course data. The main benefit of animation was that it allowed the display to be capable of conveying large amounts of information by utilising a third display dimension (i.e. time). The display also mapped time in the data to time in the display in a manner that ensured that motion could be related to meaningful qualities of the data. This allowed the users to detect and interpret a quantity of valuable patterns in their data and, specifically, find patterns of common activity over intervals of the data that could not be found using established techniques. Once a pattern was detected, the user could interact with a static frame of the animation and relate that view with complementary linked views, to read the pattern, in order to extract valuable knowledge.
A basic version of the technique was first shown at Information Visualisation 2002 in London. This demonstrated a novel display technique, which operates over a continuous temporal subset of the time series, with direct manipulation of the parameters defining the subset.
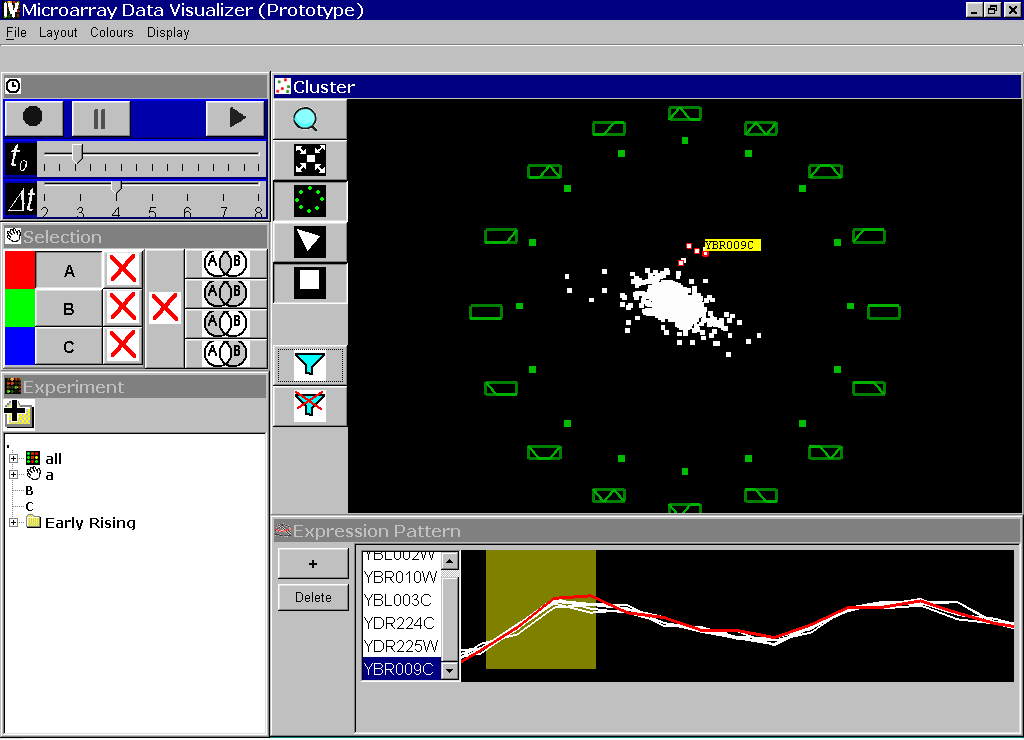
Later, we extended the technique to include a linked line-graph and sliders to define query parameters. This version of the tool was demonstrated at InfVis2003 in Seattle.
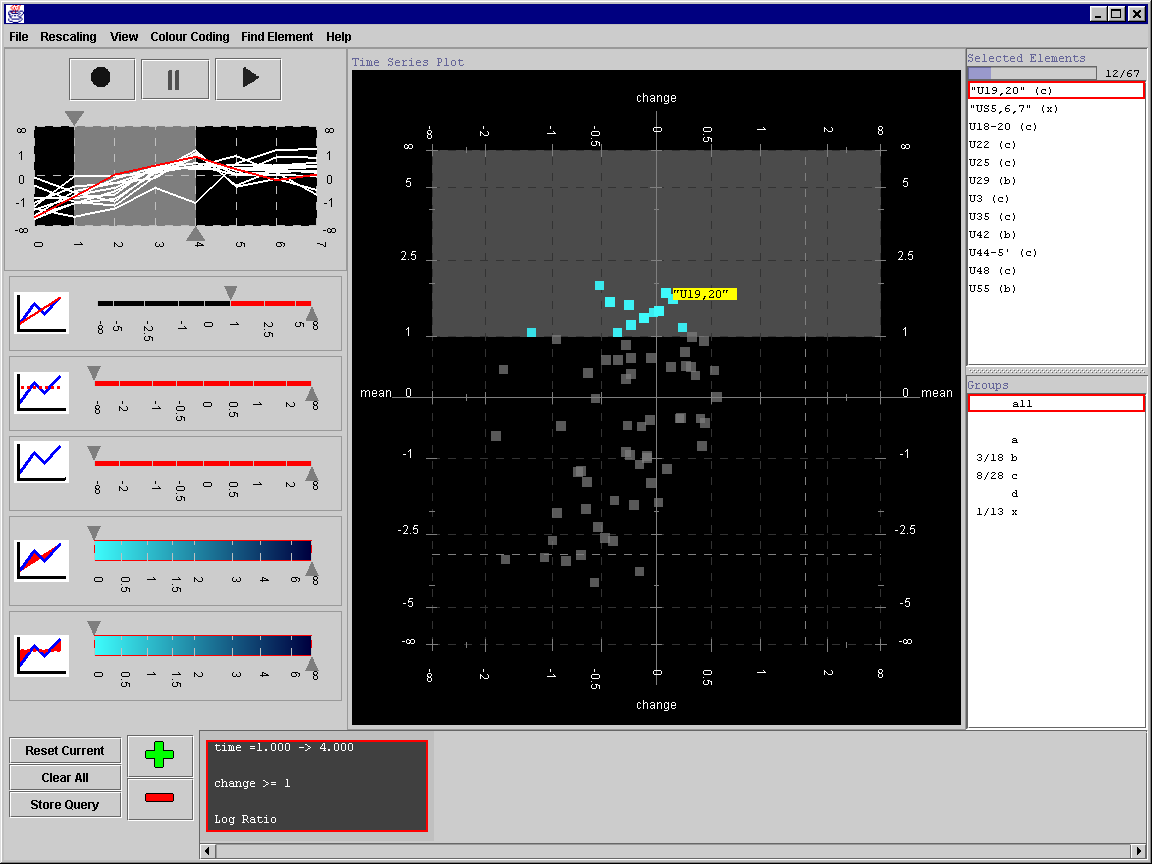
The final version of the tool allowed the user to specify queries by interacting directly with the scatter-plot. It also included features such as colour-density mapping for occluded genes and excentric labelling. We were also able to evaluate this version of the tool to find a number of expected and unexpected patterns in microarray gene expression data. These features and results were presented at CMV2005 (part of IV2005) and publishes in the Journal of Information Visualisation (2006).
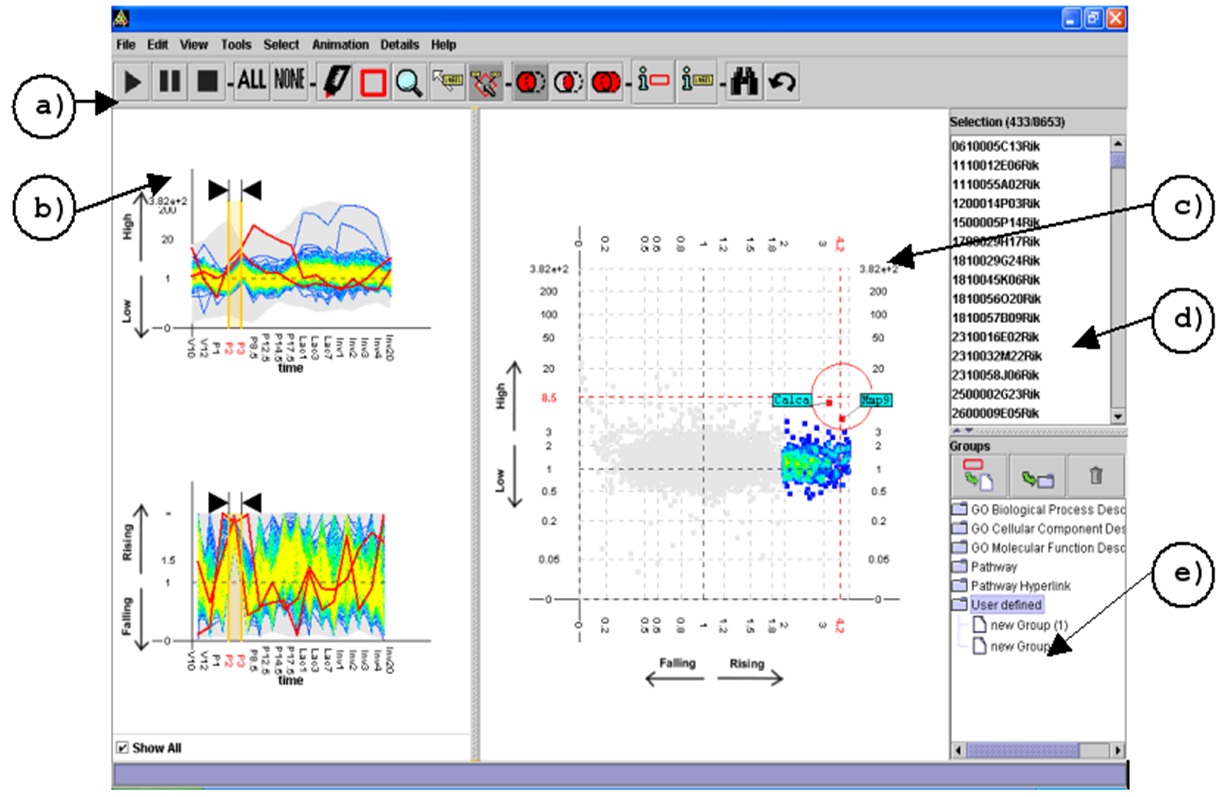
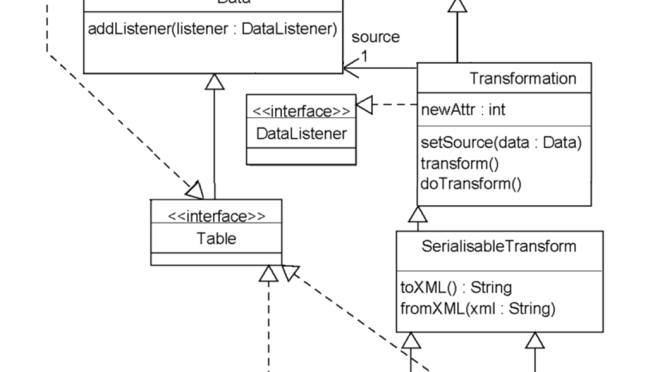
The final TSE interface
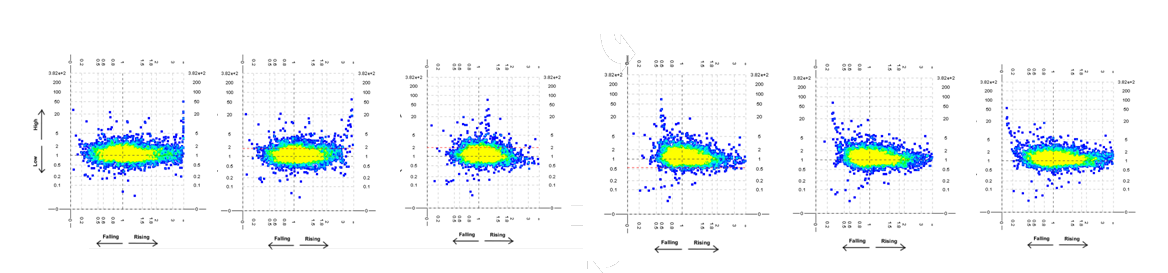 Frames from the animated scatter-plot
Frames from the animated scatter-plot
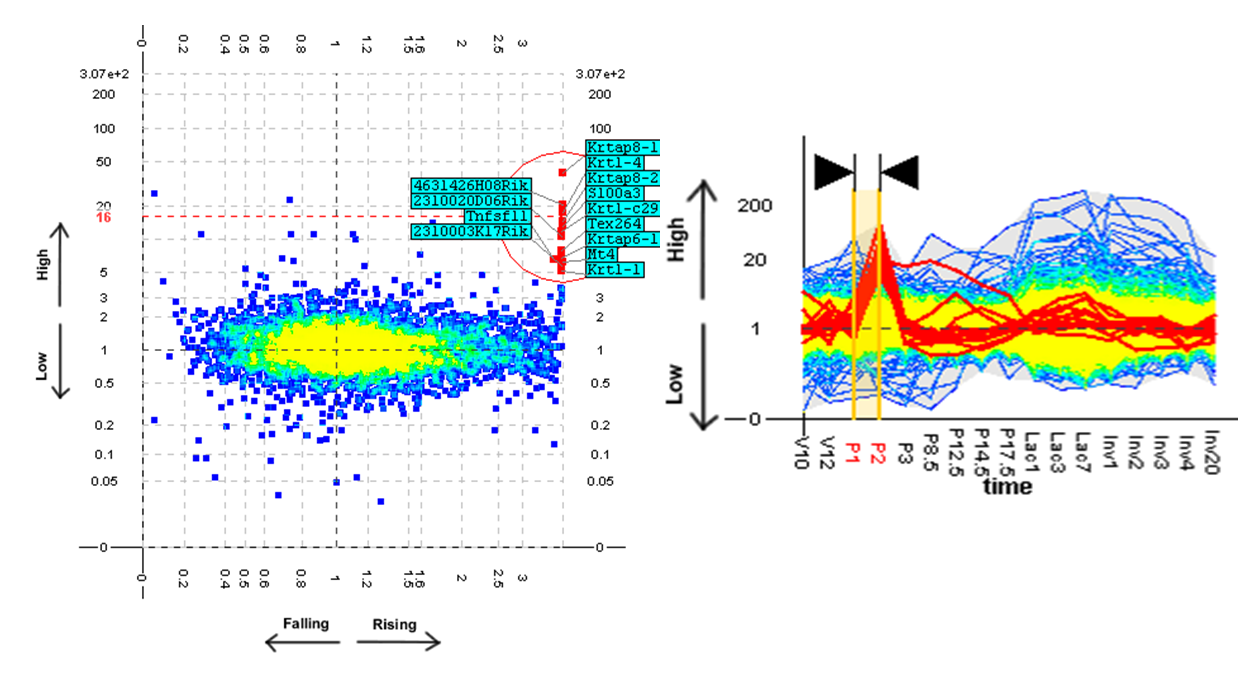 An unexpected pattern found using the TSE
An unexpected pattern found using the TSE
The results also formed an important part of my thesis (2006) and the development of TSE allowed us to prescribe a set of draft guidelines for developing animated visualisations to support the exploratory analysis of large-scale data. These are as follows:
- Animated views should be configured so that the motion has relevant meaning.
- The pace and direction of the animation should be controlled by the user.
- The motion of objects should be smoothed and regulated to avoid the undesirable effects of erratic or unpredictable motion (e.g. interpolation across time and distortion of the display space).
- Static views (coordinated and linked) should be available to help the user read a pattern once it is detected.
- Static views (coordinated and linked) can be used to help the user interpret the animation.
- The animated view can be used as a static view (see 4 and 5) when the animation is paused.
A general conclusion to be drawn from the work is that animation, where time in the data is mapped to time in the data, has enormous potential to be used to great effect in an information visualisation. While the effects of animation on human perception is less well understood than the effects of a static display, this should not discourage a developer from considering an animated visualisation if the benefits of expressiveness and an extra display dimension are required. Many of the problems with the perception of animation encountered during this project were easy to detect during user evaluations and relatively easy to eliminate after they were detected. In this case, at least, the benefits of animation were found to outstrip its disadvantages with the technique developed using animation allowing biologists to find significant patterns they could not previously find using any other techniques. It is hoped that this thesis will guide other developers toward considering the use of animation in their own information visualisation applications to achieve similar results.
Publications
Craig, P. (2006). Animated Interval Scatter-plot Views for the Exploratory Analysis of Large Scale Microarray Time-course Data (PhD). Edinburgh Napier University.
Paul Craig – thesis
Craig, P., Kennedy, J., Cumming, A. (2005). Animated Interval Scatter-plot Views for the Exploratory Analysis of Large Scale Microarray Time-course Data. Information Visualization, 4(3), 149-163.
Full colour3
Craig, P., Kennedy, J., Cumming, A. (2005). Coordinated Parallel Views for the Exploratory Analysis of Microarray Time-course Data. In: Proceedings of 3rd International Conference on Coordinated & Multiple Views in Exploratory Visualization. (pp. 3-14). London: IEEE Computer Society Press.
cmv_05
Craig, P., Kennedy, J., Cumming, A. (2005). Time-series Explorer: An Animated Information Visualisation for Microarray Time-course Data. BMC Bioinformatics 2005, 6(3), P8.
FRE Poster (A0)
Craig, P., Kennedy, J. (2003). Coordinated Graph and Scatter-Plot Views for the Visual Exploration of Microarray Time-Series Data. In: IEEE Symposium on Information Visualization. (pp. 173-180). Seattle WA: IEEE Computer Society Press.
craig
Craig, P., Kennedy, J., Cumming, A. (2002). Towards Visualising Temporal Features in Large Scale Microarray Time-series Data. In: 6th International Conference on Information Visualisation – IV2002. (pp. 427-433). IEEE Computer Society Press.
craig_microarray_02
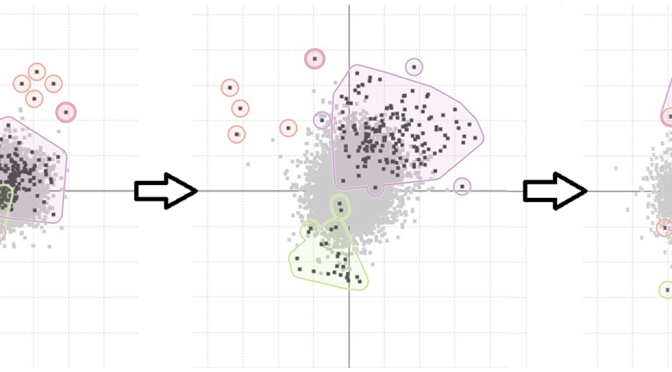
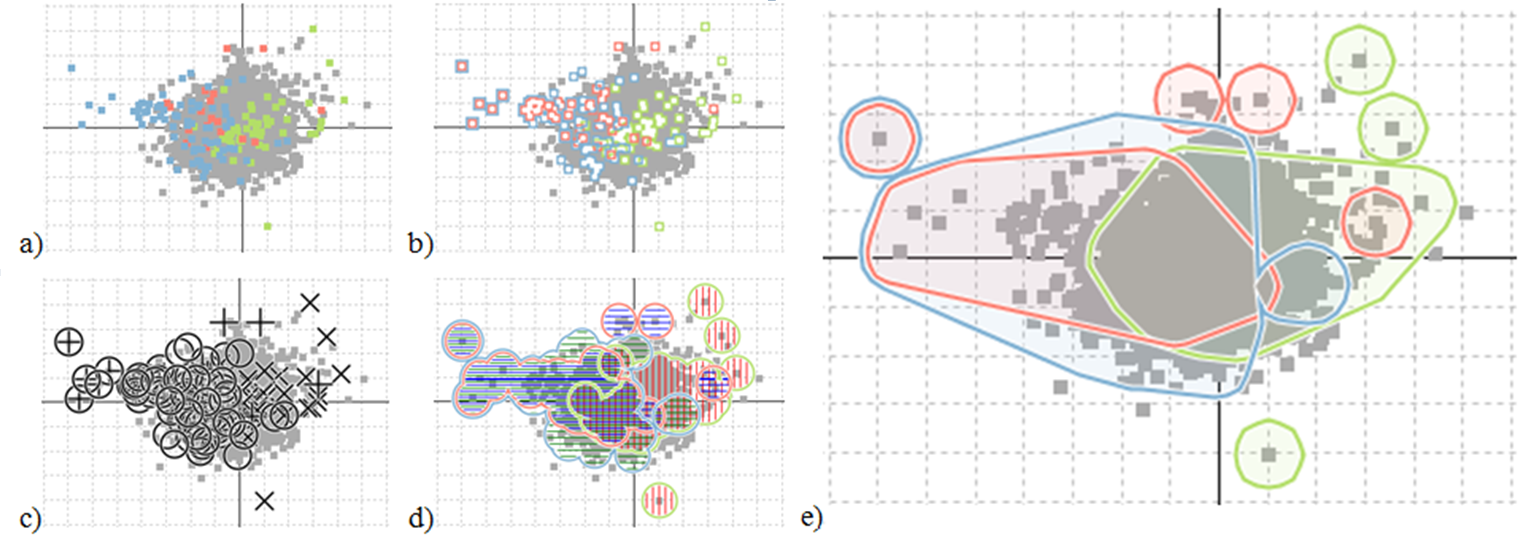
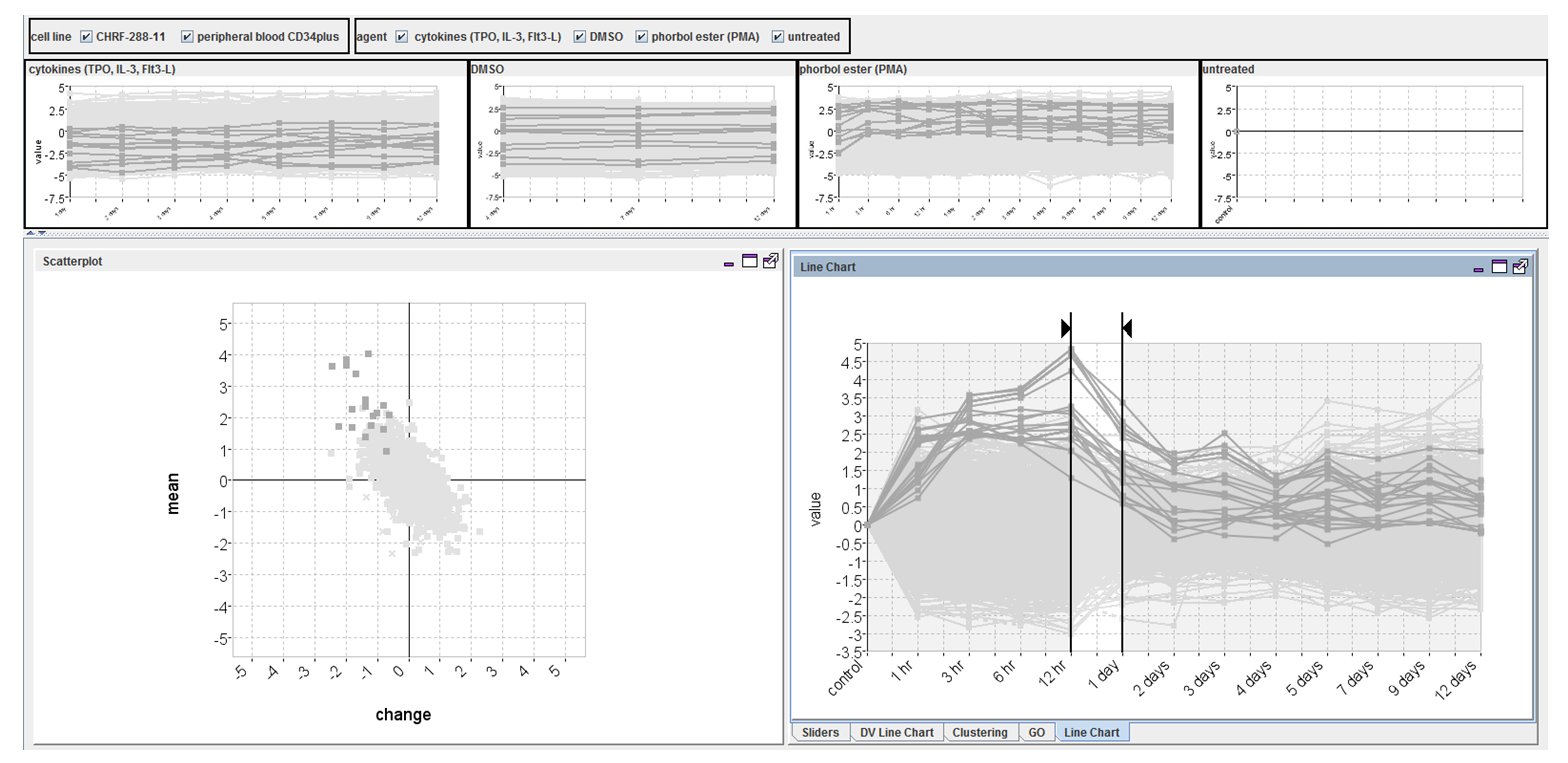
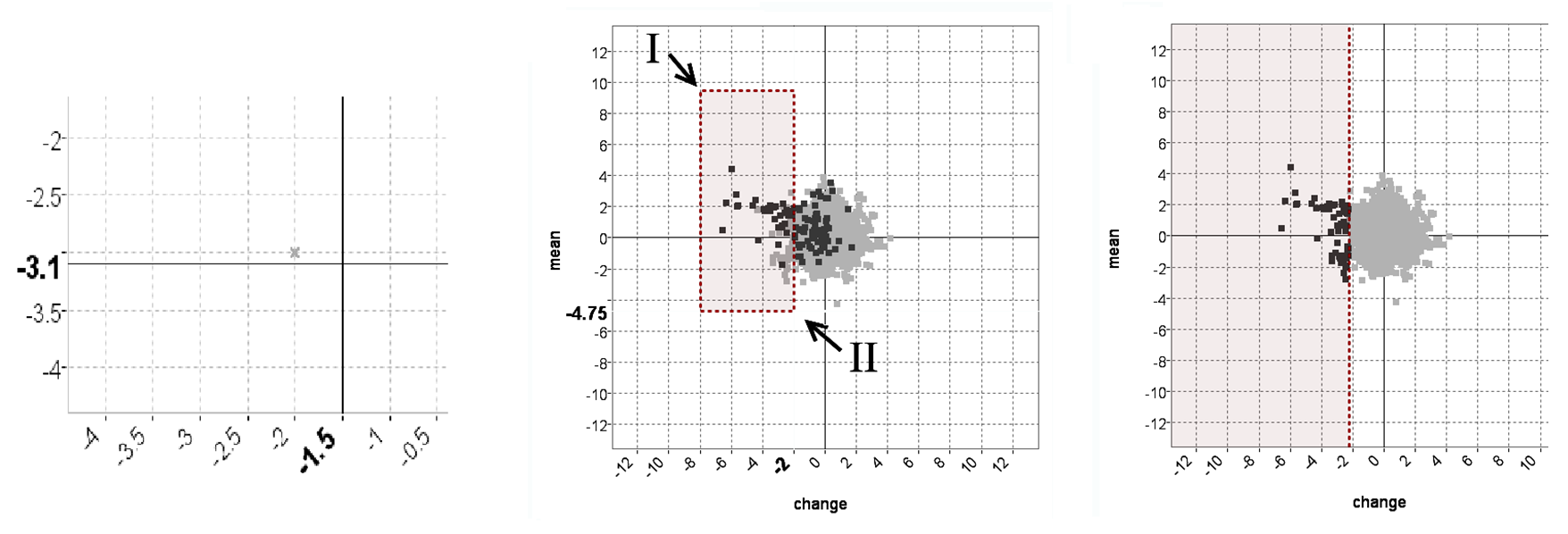
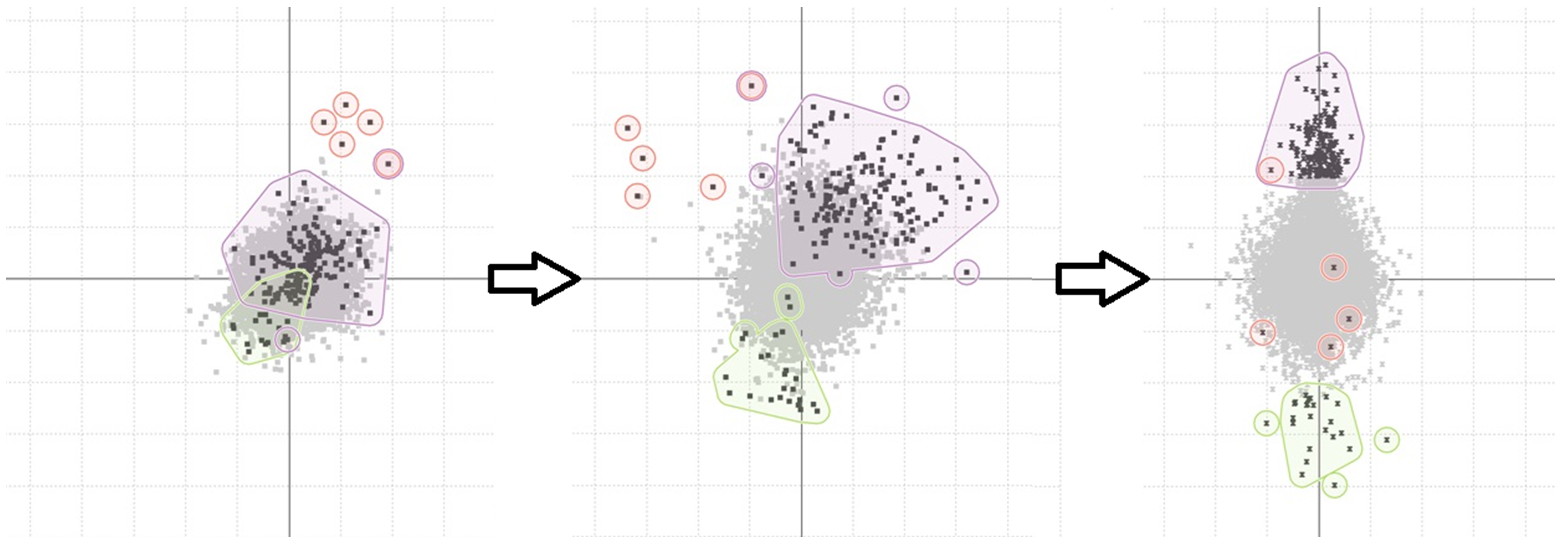
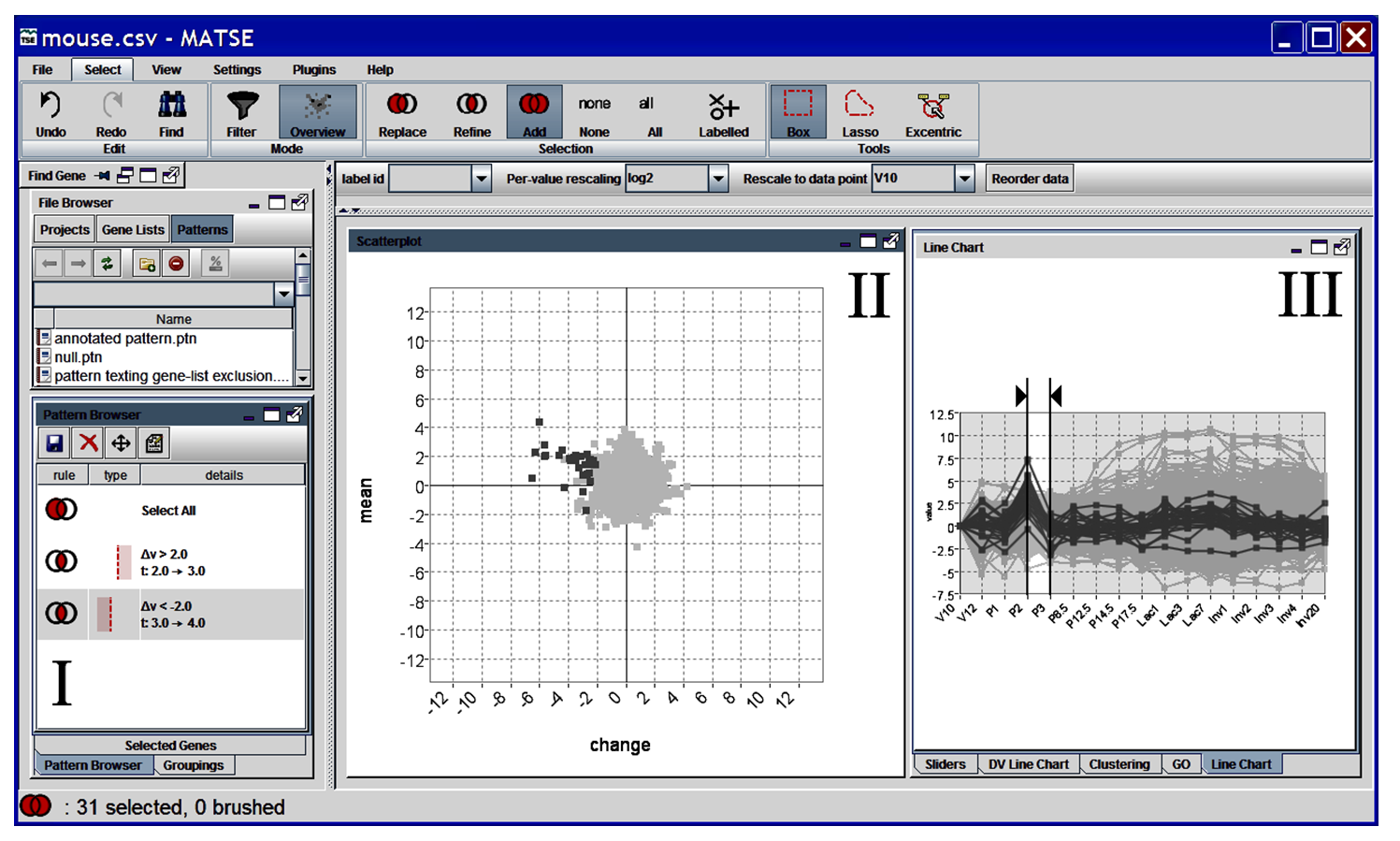
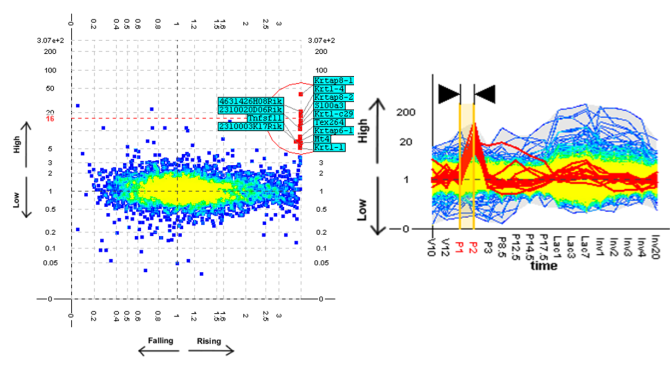




 An unexpected pattern found using the TSE
An unexpected pattern found using the TSE